Content for All Sub-Sites (Mandatory)
This is a country specific website of the Gisaid network.
Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt. At vero eos et accusam et justo duo dolores et ea rebum.
In Focus
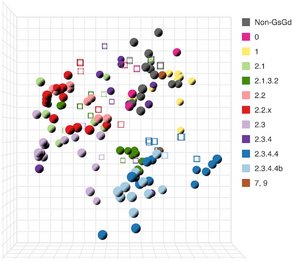
A global collaboration enabled by GISAID data sharing maps H5 influenza antigenic evolution to support vaccine development
Researchers from the University of Cambridge, the University of Wisconsin–Madison, and Erasmus Medical Center are advancing understanding of the rapid genetic and antigenic evolution of H5 influenza viruses. Using genome sequence data shared through GISAID, researchers generated synthetic hemagglutinin constructs, produced recombinant H5 viruses, and performed antigenic characterization to build comprehensive antigenic maps spanning viruses circulating since 1959. These maps guide the design of broadly protective vaccine candidates and support global surveillance.
All antigenic maps are publicly accessible and reagents are available to contributing laboratories, reinforcing the benefits from responsible data sharing, with fair attribution toward equitable global preparedness and response against emerging H5 threats
New recombinant mpox virus identified in England
Since November 2023, the Eastern parts of the Democratic Republic of the Congo (DRC) continue to see cases of the mpox virus Clade Ib that is also spreading through regional travel to neighboring countries in Central and Eastern Africa, incl. Burundi, Ethiopia, Kenya, Rwanda and Uganda. Outside of Africa, cases continue to be reported, including in Australia, Brazil, Canada, China, France, Germany, India, Italy, Japan, Netherlands, Oman, Pakistan, Portugal, Spain, Sweden, Thailand, United Kingdom and the United States.
GISAID EpiPox currently provides access to 622 genome sequences and associated metadata of the mpox virus Clade Ib. On 5 December 2025, UK Health Security Agency identified a new recombinant mpox virus which contained elements of clade Ib and IIb mpox. The specimen sample was collected from a traveller returning from Asia. The phylodynamics of Clade Ib can be monitored on GISAID’s up-to-date phylogenetic trees.
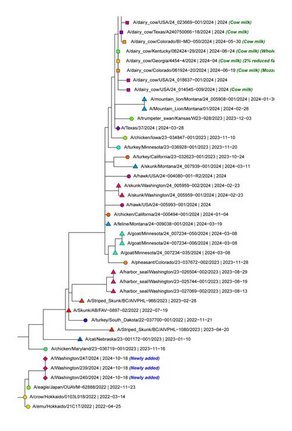
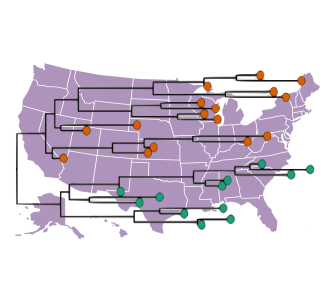
H5N1 Bird Flu continues to take its toll in the United States
Clade 2.3.4.4b of the highly pathogenic avian influenza (HPAI) virus causing outbreaks in wild and domestic birds around the world continues to spread in dairy cows, poultry and other animals across the United States. Since April 2024, the U.S. CDC confirmed 70 human cases through genome sequence analysis. Data in some of these cases showed the amino acid substitution NA-S247N known to slightly reduce susceptibility to the neuraminidase inhibitor oseltamivir in laboratory tests. In one case, a different change in the polymerase acidic (PA) protein was detected.
On 19 March 2025, the CDC released specimen data from the first human case in Ohio. The specimen sequence belongs to genotype D1.3. Previously the CDC released data from the first human fatality involving a patient from Louisiana that had been exposed to non-commercial backyard poultry and wild birds.
An Equitable Pandemic Accord?
A perspective from a Congolese Researcher
Brazzaville, Republic of the Congo - Prof. Dr. Francine Ntoumi reflects on her surveillance work in Africa during COVID-19 and shares her views on global health equity, lessons learned, the impact of technology, and the importance of international collaboration and role of GISAID's Regional Hub in Central Africa.
Dr. Francine concludes her perspective with a cautionary reminder of what a global treaty will need to provide. See also: "Data imperialism in a pandemic." On 20. May 2025, the WHO Pandemic Accord was adopted by Member States at the Seventy-eigth World Health Assembly.

GISAID joins the G20 Joint Finance & Health Ministerial Meeting
Yogyakarta, 20-21 June. Under the motto ‘Strengthening Global Health Architecture’ a delegation from GISAID attended the G20 Finance and Health Ministers meeting hosted by Indonesia, to discuss with G20 Member States’ and its partners their vision for GISAID+ (GISAID plus).
Global leaders reflected on the need to coordinate and strengthen resources during future pandemics. In particular Member States stressed the essential role GISAID plays in global health security, and discussed how global leaders could support GISAID’s expansion as a global resource to respond to other priority pathogens with major impact and pandemic-potential.

CBD Study finds 'GISAID might provide useful lessons and insight to ABS discussions'
A peer-reviewed fact-finding and scoping study on digital sequence information on genetic resources in the context of the Convention on Biological Diversity and the Nagoya Protocol, highlights key advantages of GISAID’s sharing mechanism and a fair and equitable benefit-sharing resulting from access to data.
With the core principals of timely international sharing of health data for protecting populations against lethal infectious disease outbreaks and adherence to scientific etiquette of acknowledgement of the source of data has resulted in global trust and confidence in GISAID.
Individual Content Sub-Site GisA (Optional)
Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt. At vero eos et accusam et justo duo dolores et ea rebum. Stet clita kasd gubergren. Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt. At vero eos et accusam et justo duo dolores et ea rebum. Stet clita kasd gubergren. Lorem ipsum dolor sit amet.
Mandatory Individual Content
That is content the editor of the main web-site writes for the sub-site GisA, which cannot be edited by the editor of the sub-site.
Stet clita kasd gubergren. Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt. At vero eos et accusam et justo duo dolores et ea rebum. Stet clita kasd gubergren.
GISAID Resources
![]()
Data Acknowledgement Locator

Individual Content Sub-Site (Optional)
Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt. At vero eos et accusam et justo duo dolores et ea rebum. Stet clita kasd gubergren. Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt.
At vero eos et accusam et justo duo dolores et ea rebum. Stet clita kasd gubergren. Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt.